Second-Order Effects of Chemotherapy Pharmacodynamics and Pharmacokinetics on Tumor Regression and Cachexia
by Daniel R. Bergman, Kerri-Ann Norton, Harsh Vardhan Jain & Trachette Jackson
Read the paper
This paper presents a novel computational framework that constrains high-dimensional ABM parameter space with multidimensional real-world data. We accomplish this by extending and validating a first-of-its-kind method that leverages explicitly formulated surrogate models to bridge the computational divide between ABMs and experimental data. We show that Surrogate Modeling for Reconstructing Parameter Spaces (SMoRe ParS) can constrain high-dimensional ABM parameter spaces using unidimensional (single time-course) data. We then demonstrate that it can constrain parameter spaces of more complex ABMs using multidimensional data (multiple time courses at different biological scales). To validate our method, we compared the SMoRe ParS-inferred ABM parameter space with ABM parameters inferred by an often computationally expensive direct comparison with experimental data. A strength of SMoRe ParS is that it allows for exploring ABM parameter space even at points that are not directly sampled and where ABM output was never generated. Computationally efficient methods to connect ABMs with multidimensional data are timely and important as ABMs are a natural platform for capturing heterogeneity and predicting emergent behavior in multiscale systems. SMoRe ParS is a robust and scalable computational framework that can explore the uncertainty within multidimensional parameter spaces associated with ABMs representing complex biological phenomena.
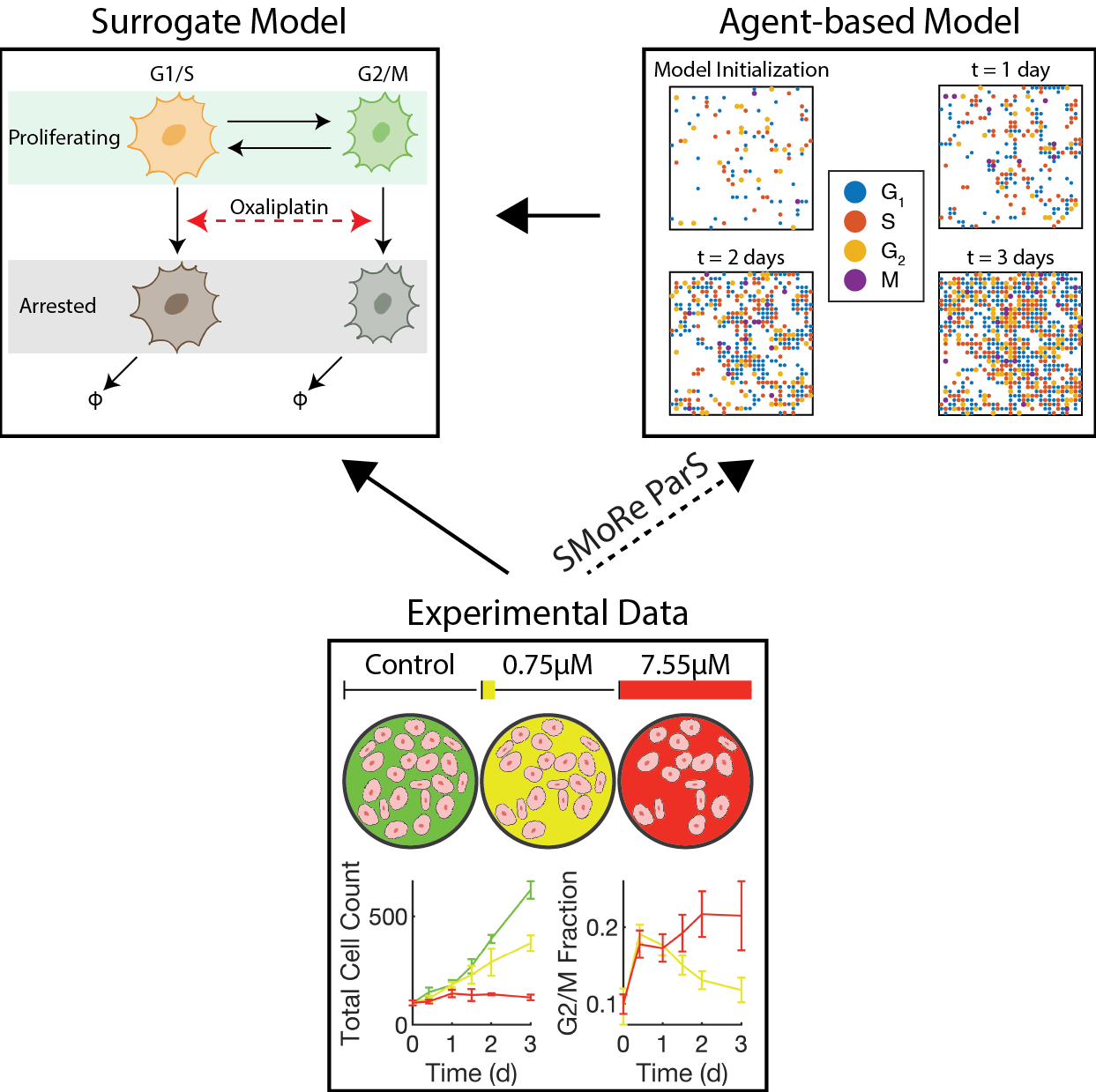
Caption: The schematic diagram for using SMoRe ParS to infer ABM input parameters from experimental data via a surrogate model. The solid arrows connecting the Experimental Data and Agent-based Model boxes to the Surrogate Model box represent the direction of information flow in the first few steps of SMoRe ParS. Green (control), yellow (0.75μM oxaliplatin), and red (7.55μM oxaliplatin) colors in the Experimental data box refer to the dosing regimens that generated the experimental data.
Brief description of the roles of the authors (e.g. student, group-leader etc):
Daniel R. Bergman, postdoc
Kerri-Ann Norton, computational modeling collaborator, and developer of SMoRe Pars
Harsh Vardhan Jain, co-senior author and co-developer of SMoRe Pars
Trachette Jackson, co-senior author and co-developer of SMoRe Pars
